Mechanobiology of Host-Microbe Interactions

Alexandre Grassart
INSERM Researcher
Alexandre's ORCID
Alexandre's Contact
The Mechano-biology of Host–Microbe Interactions team aims to understand how tissue microarchitecture, mechanical forces, and the local microenvironment modulate host–pathogen interactions at mucosal barrier interfaces. Our research primarily focuses on intestinal infections, using Shigella as a model pathogen. In this context, we develop new cell culture systems known as organs-on-chip, microphysiological systems based on microfabrication and microfluidics. Our work spans several disciplines, including cell biology, genetics, microbiology, and bioengineering.
General public
Our team studies how microbes interact with our body at sensitive surfaces such as the intestine. We aim to understand how the tissue environment and mechanical forces influence infections. In particular, we work on a bacterium called Shigella, which can cause severe diarrhea worldwide. To do this, we develop miniature models of human organs in the lab using a technology called organ-on-chip. These devices closely reproduce some functions of the intestine and allow us to observe infections in a realistic way, without using animals.
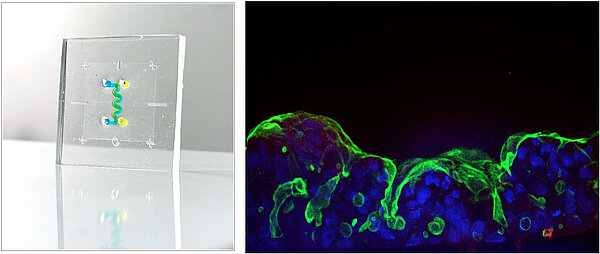
The Mechano-biology of Host–Microbe Interactions team aims to understand how tissue microarchitecture, mechanical forces, and the local microenvironment shape pathogen interactions at mucosal barrier interfaces. Our research primarily focuses on intestinal infections, using Shigella as a model pathogen. These bacteria invade epithelial cells and hijack the host intracellular machinery to propel themselves and spread rapidly to neighboring cells. This process triggers severe inflammation of the colon, leading to acute diarrhea that can be life-threatening, particularly in children under five years of age.
Like Shigella, many human pathogens are strictly human-restricted, making the study of pathophysiology at the tissue and organ levels in human-relevant contexts a major challenge. Over the past years, the team has developed a recognized expertise in the development and use of organ-on-chip (OoC) technologies for infection biology, demonstrating the high relevance of these alternative in vitro models in the field of infectiology. In particular, we have shown that Shigella infectivity is highly sensitive to mechanical forces associated with intestinal peristalsis. Building on this discovery, our goal is now to identify the strategies employed by Shigella to manipulate mechano-sensitive host pathways that promote bacterial dissemination across the epithelium.
A major axis of the lab is dedicated to continuously advancing the human relevance of in vitro models based on organs-on-chip and micro-physiological systems, combining microfabrication and microfluidics solution that can be used easily by biologists. We design complex intestinal OoC platforms that integrate key components of gut physiology, including immune cells, the enteric nervous system, inflammatory processes, and the intestinal microbiota. These systems allow us to recapitulate the dynamic, multicellular nature of the intestinal barrier under controlled mechanical and biochemical conditions.
Our research is inherently interdisciplinary, spanning cell biology, genetics, microbiology, mechanobiology, and bioengineering. In parallel, the lab is involved in numerous collaborative projects focused on the development of novel OoC models of the respiratory tract, with the aim of improving the understanding of respiratory infections and accelerating the evaluation of therapeutic strategies.
Ongoing
- France 2030-Université de Lille- Cross-Disciplinary Project: CDP MOSAIC
The interdisciplinary MOSAIC project addresses the urgent need to develop new methodological approaches (NAMs) that do not rely on animal models and better reproduce human physiology in vitro to advance translational research. MOSAIC aims to develop miniaturized devices known as organ-on-chip systems to improve our understanding of infectious and chronic diseases. This excellence-driven project brings together a consortium of 4 research units, 8 research teams, 2 hospitals, and the Lille Natural History Museum.
- France 2030 PEPR MED-OOC : projet HITOC
The HITOC project combines stem cell biology, neuroscience, infection biology, microfluidics, and microelectronics to develop a new generation of multi-segment, neuro-competent intestine-on-chip models. The goal is to better understand host–pathogen interactions and microbial signaling during viral and bacterial infections, with a particular focus on the gut–brain axis.
This project is conducted in collaboration with four additional laboratories from Inserm, CNRS, Aix-Marseille University, and Institut Pasteur.
- France 2030 PEPR TRANSCEND-ID
The main objective of this project is to establish an immunocompetent, patient-derived autologous intestine-on-chip model that reproduces the structural and functional complexity of the human intestinal mucosa and its interactions with the immune system and environment. This will enable a better understanding of chronic inflammatory bowel diseases(IBD).
This project is carried out in collaboration with two other Inserm laboratories: the INFINITE unit (Lille) and Institut Imagine (Paris).
- ANRS MIE : Projet Loc Mtb
This collaborative project with the International Center for Research in Infectiology (CIRI, Lyon) aims to better understand the earliest stages of pulmonary infection by Mycobacterium tuberculosis, the causative agent of tuberculosis. It relies on a lung-on-chip model developed in the laboratory that faithfully reproduces human alveolar physiology. This innovative approach will allow highly sensitive analysis of the infection capacity of clinical isolates and characterization of their early pathophysiological effects on human-mimetic alveolar tissue.
- Partenariat Hubert Curien Franco-Bulgare – Programme RILA
This international pilot project, conducted in collaboration with the Stephan Angeloff Institute of Microbiology (Sofia, Bulgaria), aims to better understand the early mechanisms of pulmonary inflammation leading to fibrotic interstitial lung diseases. In particular, it investigates the role of internal RNA modifications, especially m6A methylation, in amplifying pro-inflammatory immune responses, using advanced alveolar lung-on-chip models.
- MEL Accueil Talent
- ATIP-Avenir
- Start Airr Région Hauts de France
- FC3R
- Inserm First-Step
- PTR Pasteur Network PTR-430-20
Current Staff
BRODIN Priscille
INSERM research director (50%)
Priscille's ORCID
Priscille's contact
BRUGIERE Hugo
Master 2 student
Huho's contact
BURETTE Aurélie
Technician IPL
Aurélie's contact
COSTA Maxime
Post-doc Fellow
Maxime's contact
DAGAN Yoel
PhD student
Yoel's contact
DANIEL Catherine
IPL researcher
Catherine's ORCID
Catherine's contact
DEBOOSERE Nathalie
IPL Engineer
Nathalie's contact
DELANNOY Elise
Post-doc Fellow
Elise's contact
DUPRES Vincent
MCU University of Lille
Vincent's ORCID
Vincent's contact
GRASSART Alexandre
INSERM Researcher
Alexandre's ORCID
Alexandre's contact
Learnmore about Alexandre
LAFONT Frank
CNRS research director
Frank's ORCID
Frank's contact
VANPEENE Quentin
PhD student
Quentin's contact
WELLER Sandra
INSERM researcher
Sandra's ORCID
Sandra's contact
Tiffany Baetens
post-doc (2022-2024)
Grant MEL Accueil Talent
Now : Ingénieure Partenariats et innovation - Université Gustave Eiffel
Laurine de Ricker
technician (2022-2023)
Grant MEL Accueil Talent
Now : Technicienne- Laboratoire MIDAC
Zhe Ding
Visiting PhD student(2023)
Grant : First Step Inserm
Now : Post-doc UCAS (China)
Matteo Ridelfi
Visiting PhD Student (2024)
Grant : EMBO exchange
Now : Post-doc University of Maryland Baltimore (USA)
- Gut bioengineered models to study host-microbiota-probiotics interactions.
Delannoy E, Grassart A, Daniel C. Microbiome Res Rep. 2025 Oct 22;4(4):39. doi: 10.20517/mrr.2025.45. eCollection 2025. - Gut-on-chip methodology based on 3D-printed molds: a cost-effective and accessible approach.
Delannoy E, Burette A, Janel S, Poiret S, Deboosere N, Daniel C, Grassart A. Lab Chip. 2025 Aug 19;25(17):4396-4409. doi: 10.1039/d5lc00147a. - A multifunctional anti-O-Antigen human monoclonal antibody protects against Shigella sonnei infection in vivo.
Ridelfi M, Vezzani G, Roscioli E, Batani G, Boero E, Nannini F, Marini E, Bhaumik U, Desalegn G, Serpino O, Molinaro A, Paciello I, Mugnaini C, De Santi C, Maccari G, De Rosa A, Valensin S, Duatti A, Di Benedetto R, Raso MM, Alfini R, Sammicheli C, Tavarini S, Quigley C, Podda A, Kabanova A, Andreano E, Cardamone D, Gasperini G, Necchi F, Pizza M, Martin LB, Frenck RW Jr, Grassart A, Silipo A, Pasetti MF, Berlanda Scorza F, Giannelli C, Rossi O, Micoli F, Sala C, Rappuoli R. Proc Natl Acad Sci U S A. 2025 Jul 29;122(30):e2426211122. doi: 10.1073/pnas.2426211122. Epub 2025 Jul 23. - Organoids, organs-on-chips, complex in vitro model: Definitions, applications, validation, ethics.
Mottet G, Grassart A, Barthélemy P, Antignac C, Arrabal S, Bourdin A, Descroix S, De Vos J, Doutriaux A, Fabrega Q, Galaup A, Graff-Dubois S, Illiano S, Legallais C, Maisonneuve B, Piwnica D, Quéméneur E, Salentey V, Rozenberg J, Sotiropoulos A, Tomasi R, Vergnolle N, Devillier P. Therapie. 2025 Jan-Feb;80(1):17-31. doi: 10.1016/j.therap.2024.12.002. Epub 2024 Dec 4. - 4D live imaging and computational modeling of a functional gut-on-a-chip evaluate how peristalsis facilitates enteric pathogen invasion.
Boquet-Pujadas A, Feaugas T, Petracchini A, Grassart A, Mary H, Manich M, Gobaa S, Olivo-Marin JC, Sauvonnet N, Labruyère E. Sci Adv. 2022 Oct 21;8(42):eabo5767. doi: 10.1126/sciadv.abo5767. Epub 2022 Oct 21. - Bioengineered Human Organ-on-Chip Reveals Intestinal Microenvironment and Mechanical Forces Impacting Shigella Infection.
Grassart A, Malardé V, Gobaa S, Sartori-Rupp A, Kerns J, Karalis K, Marteyn B, Sansonetti P, Sauvonnet N. Cell Host Microbe. 2019 Oct 9;26(4):565. doi: 10.1016/j.chom.2019.09.007. - Shigella promotes major alteration of gut epithelial physiology and tissue invasion by shutting off host intracellular transport.
Ferrari ML, Malardé V, Grassart A, Salavessa L, Nigro G, Descorps-Declere S, Rohde JR, Schnupf P, Masson V, Arras G, Loew D, Sansonetti PJ, Sauvonnet N. Proc Natl Acad Sci U S A. 2019 Jul 2;116(27):13582-13591. doi: 10.1073/pnas.1902922116. Epub 2019 Jun 17.





