Molecular & cellular virology of hepatitis E virus
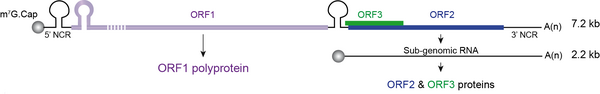
Hepatitis E virus (HEV) is the major cause of acute viral hepatitis worldwide. Although infection with HEV is usually self-resolving, it can cause up to 30% mortality in pregnant women and leads to a chronic infection in immunocompromised patients. There is no specific treatment nor universal vaccine to fight against this virus. HEV is a quasi-enveloped, positive-sense RNA virus expressing three open reading frames (ORFs): ORF1 encodes the ORF1 viral replicase, ORF2 encodes the ORF2 viral capsid protein and ORF3 encodes a small protein involved in virion morphogenesis and egress. Due to poor replication in cell culture, many data are missing on the different steps of the HEV lifecycle. In our group, we are mainly studying the replication and assembly mechanisms of HEV, and the biogenesis of ORF2 capsid protein. We are also developing tools to probe viral proteins. Besides a better understanding of the HEV life cycle, our research program aims to identify novel antiviral targets.
Development of a HEV cell culture system and identification of different forms of the ORF2 capsid protein
Few years ago, we developed an efficient cell culture system for HEV which allows for studying the molecular and cellular mechanisms of the HEV lifecycle. This model notably enabled the pioneering demonstration that, during its lifecycle, HEV produces at least 3 forms of the ORF2 capsid protein: infectious ORF2 (ORF2i), glycosylated ORF2 (ORF2g), and cleaved ORF2 (ORF2c). These ORF2 forms perform distinct functions in the HEV lifecycle. The ORF2i protein is the structural component of infectious particles whereas the ORF2g/c proteins are not associated with infectious material but likely act as a humoral immune decoy that inhibits antibody-mediated neutralization. The ORF2g/c proteins are secreted in large amounts in culture supernatant and are the most abundant antigens detected in patient sera.
We are currently investigating whether other ORF2 forms are produced in vitro and in vivo.
doi: 10.1053/j.gastro.2017.09.020
doi: 10.1038/s41598-019-42737-2
Identification and characterization of HEV factories
We generated monoclonal antibodies that specifically recognize the particle-associated ORF2i form, and antibodies that recognize the different ORF2 isoforms. We used them in confocal and electron microscopy approaches to probe infectious particles and viral factories in HEV-producing cells. We identified an unprecedented HEV-induced membrane network containing tubular and vesicular structures that were enriched in ORF2 and ORF3 viral proteins. Moreover, extensive colocalization analyses of viral proteins with subcellular markers, such as Rab11, demonstrated that these structures were derived from the endocytic recycling compartment (ERC). In addition, in a study aiming at characterizing the ORF1 replicase and using the RNAscope technology, we revealed that the ORF1 protein and genomic RNA were also enriched in the ERC-derived subcellular structures, indicating that cellular ERC serves a viral factory for HEV. Thus, HEV hijacks the ERC during its lifecycle and forms a membrane network of vesicular and tubular structures that might be the hallmark of HEV infection.
Very recently, we demonstrated that the AP-1 adaptor complex plays a pivotal role in the targeting of ORF2i protein to viral factories. This complex belongs to the family of adaptor proteins that are involved in vesicular transport between the trans-Golgi network and early/recycling endosomes. We demonstrated that the ORF2i protein colocalizes and interacts with the AP-1 adaptor complex in HEV-producing or infected cells. We showed that silencing or drug-inhibition of the AP-1 complex prevents ORF2i protein localization in viral factories and reduces viral production in hepatocytes. Modeling of the ORF2i/AP-1 complex also revealed that the S domain of ORF2i likely interacts with the σ1 subunit of AP-1 complex. Hence, our study identified for the first time a host factor involved in addressing HEV proteins (i.e. ORF2i protein) to viral factories.
Our current projects aim to identify other cellular and viral effectors involved in the formation of viral factories.
doi: 10.1007/s00018-022-04646-y
doi: 10.3389/fmicb.2022.828636
doi: 10.21203/rs.3.rs-4539560/v1
HEV replication
Knowledge on the replication mechanisms of HEV remains scarce. The viral replicase is encoded by ORF1 and is composed of several functional domains. During the replication process, a subgenomic RNA is produced to give rise to ORF2 and ORF3 proteins. The viral determinants and the cellular environment or factors necessary for the optimal HEV replication remains largely to be explored. Thanks to our efficient culture system to study the entire HEV lifecycle and established replicons to specifically study HEV replication, we identified the guanine exchange factor GBF1 as a cellular factor acting in HEV replication. Our current projects aim at identifying and characterizing other effectors of HEV replication as well as identifying plant-derived drugs targeting HEV replication.
doi: 10.1111/cmi.12804
Biogenesis of the ORF2 isoforms
Recently, we deciphered the mechanisms by which the ORF2 forms are produced and differentially addressed to cell compartments. We demonstrated that HEV has set up a nucleo-cytoplasmic transport mechanism of its capsid protein to modulate cell host immune responses. In addition, we found that during the HEV lifecycle, a fine-tuning of ORF2 partitioning occurs between cytosolic, reticular and nuclear compartments. Importantly, we identified a stretch of 5 amino acid residues (named ARM, Arginine-Rich Motif) in the N-terminal region of the ORF2 protein that drives nuclear translocation and tightly modulates the stoichiometry between the different ORF2 forms, especially by regulating the functionality of the ORF2 signal peptide and interactions with the host cell. Thus, the ORF2 ARM is a unique central regulator of ORF2 addressing that finely controls the HEV lifecycle.
We are currently continuing to characterize the biogenesis pathways of ORF2 forms and the post-translational modifications they undergo.
doi: 10.1371/journal.ppat.1010798